Urban Sound Classification using Machine Learning: Experiments
Cross validation
When learning machine learning algorithms, we were taught using train_test_split() method in sklearn.model_selection, which splits samples into random train and test subsets. If we apply this method to our samples, which were constructed from all folders(fold1~fold10) in the dataset, the rows of the sample are shuffled in random selections. The authors mentioned 'do not shuffle' because
"If you reshuffle the data (e.g. combine the data from all folds and generate a random train/test split) you will be incorrectly placing related samples in both the train and test sets, leading to inflated scores that don't represent your model's performance on unseen data. Put simply, your results will be wrong.""
Overfitting
The early stage of experiments shows the following training history with neuron=128, showing overfitting:
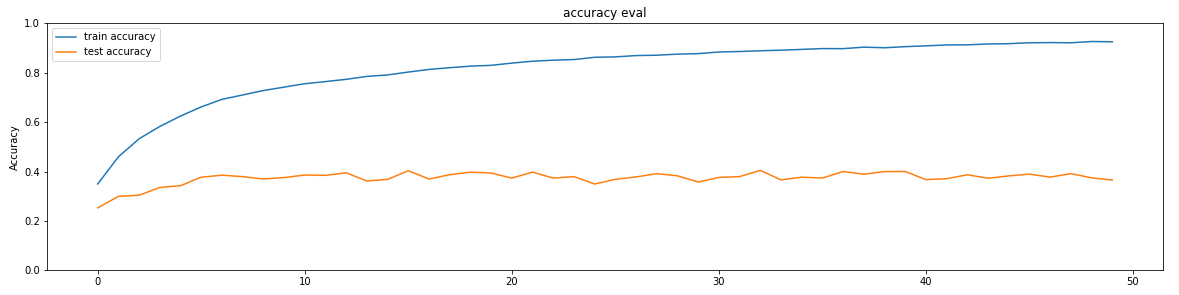 To avoid the overfitting, first we used cross validation as in shown in the
below table. Secondly, we drop the
training accuracy to obtain better test accuracy by using dropout, leading the line
graphs get closer.
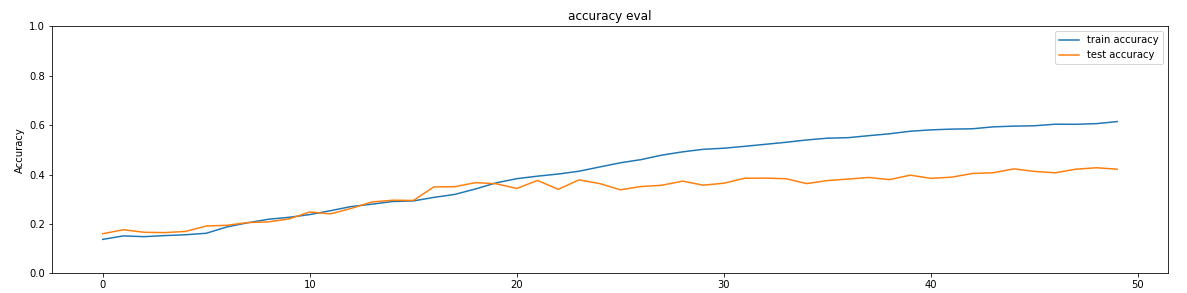
To avoid the overfitting, first we used cross validation as in shown in the
below table. Secondly, we drop the
training accuracy to obtain better test accuracy by using dropout, leading the line
graphs get closer.
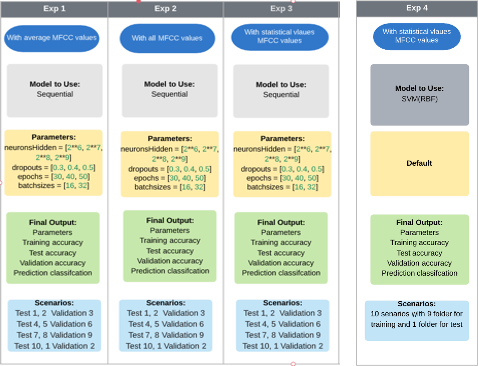
Experiments
Experiment 1, 2 and 3
- number of neurons = [64, 128, 256, 512]
- dropouts = [0.3, 0.4, 0.5]
- epochs = [30, 40, 50]
- batch sizes = [16, 32]
| Folders for test | Folders for validation | Folders for train |
|---|---|---|
| fold 1, 2 | fold 3 | fold 4, 5, 6, 7, 8, 9, 10 |
| fold 4, 5 | fold 6 | fold 7, 8, 9, 10, 1, 2, 3 |
| fold 7, 8 | fold 9 | fold 10, 1, 2, 3, 4, 5, 6 |
| fold 10, 1 | fold 2 | fold 3, 4, 5, 6, 7, 8, 9 |
Experiment 4
Experiment Scenarios diagram
Eunjeong Lee, ejlee127 at gmail dot com, last updated in Nov. 2020